Dimitar Kostadinov
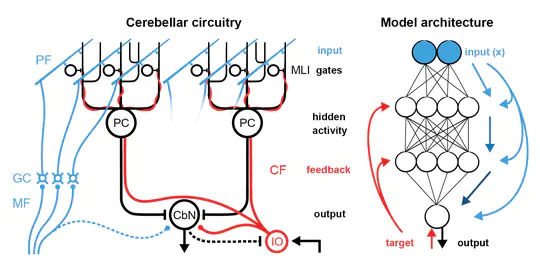
Next spring, I will be starting my own lab in the Centre for Developmental Neurobiology at King’s College London. The lab is going to study how the cerebellum contributes to multiregional neural computations for behavioural execution and learning by combining functional, computational, and molecular approaches. I am looking to hire at all levels, so please get in touch if you are interested.
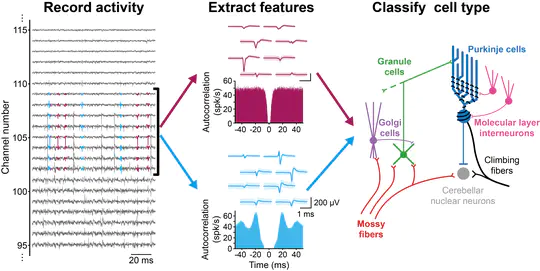
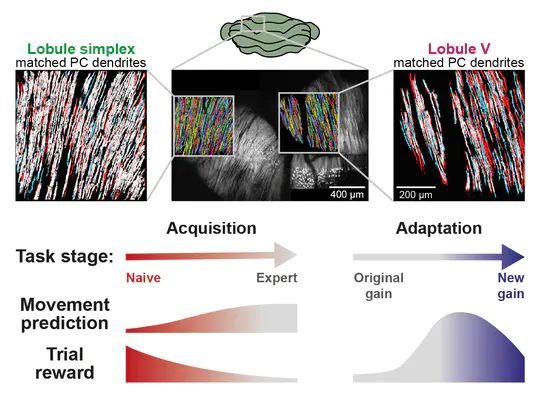
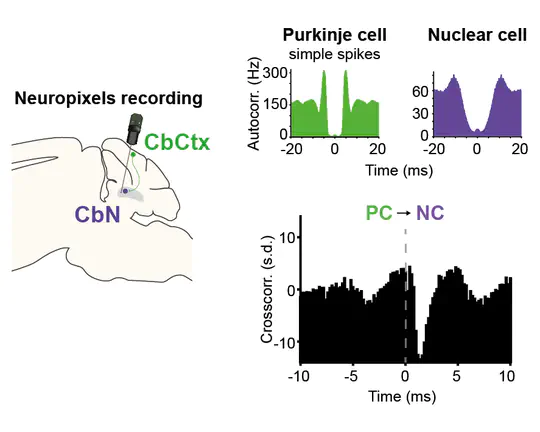
I am currently a postdoctoral fellow in Michael Häusser’s Neural Computation Laboratory in the Wolfson Institute for Biomedical Research at University College London. Over the last several years, I have been studying cerebellar computation during goal-directed behaviour using two-photon imaging, high-density electrophysiology, and optogenetics.
Prior to this, I was a PhD student in the Program in Neuroscience at Harvard University with Josh Sanes. I studied the mechanism and function of dendritic self-avoidance in the mammalian retina using a combination of mouse genetics, anatomy, and patch-clamp electrophysiology.
Currently reading: Some Rain Must Fall: My Struggle 5 by Karl Ove Knausgård.
Currently listening to: Wet Leg by Wet Leg.
Ongoing projects
Publications
Resources
A collection of causes and topics of interest